Advertisements
Advertisements
प्रश्न
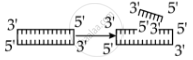
Identify the activity of endonuclease and exonuclease in the given image.
विकल्प
Endonuclease Exonuclease 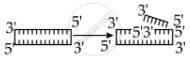
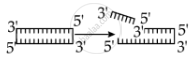
Endonuclease Exonuclease 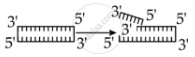
Endonuclease Exonuclease 
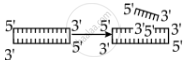
Endonuclease Exonuclease 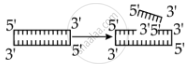

उत्तर
| Endonuclease | Exonuclease |
 |
 |
Explanation:
Endonucleases are a type of restriction enzyme that cuts the DNA at a specified location. It is widely used to clip the sequence in order to obtain DNA fragments with sticky ends. Sticky ends are single-stranded DNA segments that can establish hydrogen bonds with adjacent cut DNA segments. The enzyme ligase later joins these ends. Exonuclease is a restriction enzyme that removes nucleotides from the 5' and 3' ends of DNA strands.
APPEARS IN
संबंधित प्रश्न
Make a chart (with diagrammatic representation) showing a restriction enzyme, the substrate DNA on which it acts, the site at which it cuts DNA and the product it produces.
Do eukaryotic cells have restriction endonucleases? Justify your answer.
Give a reason why :
Single cloning site is preferred in a vector.
The total number of nucleotide sequences of DNA that code for a hormone is 1530. The proportion of different bases in the sequence is found to be Adenine = 34%, Guanine = 19%, Cytosine = 23%, Thymine = 19%.
Applying Chargaff’s rule, what conclusion can be drawn?
Molecular scissors, which cut DNA at specific site is ______.
DNA strands on a gel stained with ethidium bromide when viewed under UV radiation, appear as ______
Which of the given statements is correct in the context of visualizing DNA molecules separated by agarose gel electrophoresis?
A plasmid DNA and a linear DNA (both are of the same size) have one site for a restriction endonuclease. When cut and separated on agarose gel electrophoresis, plasmid shows one DNA band while linear DNA shows two fragments. Explain.
Carefully observe the given picture. A mixture of DNA with fragments ranging from 200 base pairs to 2500 base pairs was electrophoresed on agarose gel with the following arrangement.
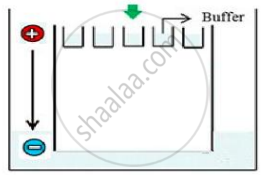
(a) What result will be obtained on staining with ethidium bromide? Explain with reason.
(b) The above setup was modified and a band with 250 base pairs was obtained at X.
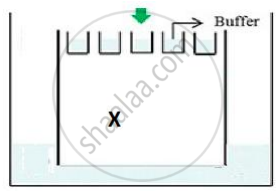
What change(s) were made to the previous design to obtain a band at X? Why did the band appear at position X?
Given below is the restriction site of a restriction endonuclease Pst-I and the cleavage sites on a DNA molecule.
\[\ce{5' C - T - G - C - A \overset{\downarrow}{-}{G 3'}}\]
\[\ce{3' G\underset{\uparrow}{-} A - C - G - T - C 5'}\]
Choose the option that gives the correct resultant fragments by the action of the enzyme Pst-I.
