Advertisements
Advertisements
Question
Do eukaryotic cells have restriction endonucleases? Justify your answer.
Solution
Yes, restriction endonucleases are found in eukaryotic cells. Restriction endonuclease acts by examining the length of the DNA sequence. When it finds its specific recognition sequence, it binds to the DNA and cuts both strands of the double helix at specific points in the sugar-phosphate backbones. Each restriction endonuclease recognises specific palindromic nucleotide sequences in the DNA.
APPEARS IN
RELATED QUESTIONS
Explain with the help of a suitable example the naming of a restriction endonuclease.
Mention the difference in the mode of action of exonuclease and endonuclease.
Name the enzymes that are used for the isolation of DNA from bacterial and fungal cells for recombinant DNA technology.
How does a restriction nuclease function? Explain
Collect 5 examples of palindromic DNA sequences. Better try to create a palindromic sequence by following base-pair rules.
How does restriction endonuclease function?
The total number of nucleotide sequences of DNA that code for a hormone is 1530. The proportion of different bases in the sequence is found to be Adenine = 34%, Guanine = 19%, Cytosine = 23%, Thymine = 19%.
Applying Chargaff’s rule, what conclusion can be drawn?
The DNA fragment separated on an agarose gel can be visualized by staining with ______.
A mixture containing DNA fragments a, b, c and d, with molecular weights of a + b = c, a > b and d > c was subject to agarose get electrophoresis. This position of these fragments from cathode to anode to anode sides of the gel would be ______.
Which of the following radioisotope is not suitable for DNA labeling based studies?
A specific recognition sequence identified by endonucleases to make cuts at specific positions within the DNA is ______
While isolating DNA from bacteria, which of the following enzymes is not required?
Would you choose an exonuclease while producing a recombinant DNA molecule?
How does one visualise DNA on an agarose gel?
CTTAAG
GAATTC
- What are such sequences called? Name the enzyme used that recognizes such nucleotide sequences.
- What is their significance in biotechnology?
Carefully observe the given picture. A mixture of DNA with fragments ranging from 200 base pairs to 2500 base pairs was electrophoresed on agarose gel with the following arrangement.
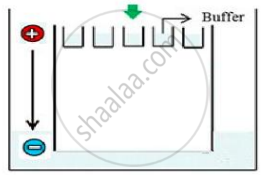
(a) What result will be obtained on staining with ethidium bromide? Explain with reason.
(b) The above setup was modified and a band with 250 base pairs was obtained at X.
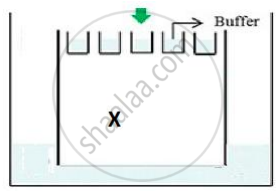
What change(s) were made to the previous design to obtain a band at X? Why did the band appear at position X?
'EcoRI' has played a very significant role in rDNA technology.
- Explain the convention for naming EcoRI.
- Write the recognition site and the cleavage sites of this restriction endonuclease.
